Science method
Staining - Science method
Explore the latest questions and answers in Staining, and find Staining experts.
Questions related to Staining
I'm now doing western blotting for several proteins.
but transfer is not perfectly happened.
bands are looks like smeared or smudged.
And it seems to the lower MW bands are more affected.
what did I do wrong?
I use towbin buffer without SDS / PVDF 0.2um pore / biorad wet blot tank transfer system / during transfer, use stirring and ice block with 400mA (about 200~100V)
Also, when I performed with 20V overnight same thing happened.
I attached picture of my transferred membrane
when I stained membrane with Ponceau S solution, other protein bands looks same as marker band.

I would like to know if anyone tried the western blot with the Instant Blue stained SDS-PAGE gels. Thanks.
Hello everyone,
Recently I've been working on the immunofluoresence of HepG2 cell lines. I tried both DAPI (1µg/ml, in PBS, 30mins RT) and mounting medium with DAPI but the cell membrane was always stained by it. Pics are attached below. Does anyone know the reason? I would be really appreciated for your time!
Hello everyone,
I want to stain live and dead cells in a fresh tissue. There are a large variety of stains (i.e, PI, cell tracker, FDA, DAPI). As long as I know, the should not be exposed to light. The pathology facility I work with, does not have a dark environment( the samples would be exposed to light). Can you recommend a staining agent that does not degrade in light exposure?
I did an immunofluorescent experiment with both unstimulated and stimulated MC/9 cells and stained for CD107a. I didn't permeabilise the cells, however, I noticed strong intracellular staining in unstimulated cells (CD107a is inside the cells, so I shouldn't have seen any staining). Does anyone know what happened?
I am working on Exosomes that is derived from cells that are engineered to express scFv of Anti-HER2 antibody. I want to see its expression in TEM. Can I directly label the exosomes with the Protein L-GNP or do I need to use Anti-Anti HER2 antibody and secondary antibody then stain with Protein L GNP?
Can anyone suggest literature on Protein L-GNP immuno gold staining in TEM?
I would like to track in real time the growth of bacteria using fluorescent miscoscopy. I remember there was at least one plasmid that could be used: transfected bacteria would emit a red or green fluorescent signal that could be used to track their growth and position.
Alas, I don't remember what was the name of these plasmids and I can't find a reference in the literature.
Does somebody know these kind of plasmids for live tracking of bacteria? Where can I buy them?
Thank you.
Hi everyone,
I am staining leukocytes isolated from mouse kidneys in flow cytometry. I stimulated the cells with PMA/Ionomycin + BFA for 4 hrs in T-cell medium (RPMI + Pen/Strep + 10 % FCS + 50 uM beta-mercaptoethanol) and stained them for surface markers, T-cell transcription factors and IFNg and IL17A.
When I plot the cytokine production (IFNg-BV711 vs IL17A-BV650) in CD4+ T cells, I noticed that beside my single-positive populations, there are events on a somewhat straight diagonal line that seem to be double-positive. There are some other events that are double-positive that are more scattered around, which is why I think the events on the diagonal could be a technical artifact (see attached plot).
I am also attaching my FMOs for IFNg-BV711 and IL17A-BV650 where these events are not present.
I'd highly appreciate your thoughts on this.
Thanks a lot,
Jasper



I want to stain acute brain slices with calcium orange and DAPI, but I am not sure whether these two kinds of dye can be added together or not.
I am planning on staining neuronal cells in culture to visualize neurites. Thermofisher's Vybrant-CM DiI seems the best fit for my requirements. However, I am unable to find any images or papers that show the use of the this stain on neuronal cells. I am now reconsidering the usage of this stain, and am also considering the Vybrant DiI solution or the DiI crystals also sold by Thermofisher. If you have used this with neurons, please comment on it!
NOTE: I aim to use a non-injectable stain.
Hello,
I am planning to conduct an experiment to identify bacteria bound to IgG by flow cytometry. I aim to focus on live bacteria, so I intend to use the LIVE/DEAD BacLight Kit (Syto9 and propidium iodide) to confirm I'm examining live bacteria. Since I need to fix the samples before acquisition on the flow cytometer, my questions are:
- Is it possible to fix samples when using the LIVE/DEAD BacLight Kit?
- Should I perform IgG staining before or after the LIVE/DEAD staining?
Thanks in advance,
I try to mark Proximal tubular cells with LTL while doing immunofluorescence staining.
I use BioWorld LTL-Texas Red. I tried 1:100 for 1 h after secondary antibody and 1:50 overnight with primary antibody. Both conditions didn't give good staining. any suggestions?
Hi everyone! I have carried out SDS PAGE analysis many times. In the experiment, protein sample was hydrolyzed gelatin. After staining, only bands of protein marker (standard) were shown, whereas sample bands were unclear or invisible. Is a scanner always used? If somebody knows please give me some pieces of advice. Many thanks for considering my request.
I am staining mouse brain tissue sections using an anti-GFAP primary antibody (for astrocytes). The results are coming out pretty weird. Some sections have decent staining, and some others have horrible/very weak staining. I'm using the same protocol and reagents as I usually do (and have had successful staining in the past), so I won't go into those details here.
Instead, this was my first time transcardially perfusing the animals with paraformaldehyde. I suspect some of the perfusions did not go well: a couple of the bodies did not get very stiff. After brain extractions, I put them in a PFA/sucrose solution overnight. Looking at my stained sections under the microscope, the clearance of blood didn't seem to be a huge issue. There is a little autofluorescence going on (due to the blood, I suspect), but overall, there is no blood in the sections.
So, could it be that the PFA didn't penetrate my tissue well? Would this cause extremely weak signal in my sections (even though the tissue did sit in PFA overnight)?
The pictures are examples.
1) The staining came out as expected on this one.
2) Verrrry weak signal, but you can see the GFAP.
3) An example of autofluorescence from the blood that was left. I see absolutely no GFAP/astrocytes.



I am conducting the IHC experiment with cultured cells.
However, I experienced the non-specific staining with all my samples.
The nucleus were stained by DAPI, but beside the blue of DAPI, they were also stained by the conjugated red-fluorescent dye of secondary antibody. I noticed that the cell nucleus was stained even much more strongly red than the other target parts. So when I merged two color images together, the red and blue signals overlapped, causing them to turn purple-pink. How to remove non-specific red staining of cell nucleus?
I put my image and our protocol below.
1. Fixation step: 4% Paraformaldehyde (PFA) 2ml, 20 min, Room Temp. --> 1. Washing with PBS (shaking) (3 times)
2. Permeation: 0.1% Triton in PBS for 10 min --> Washing with PBS (5min)
3. Blocking: 10% Serum in PBS, 1.5h
4. Primary antibody: Remove blocking solution, not washing --> Incubate cells in working solution of (0.1% Triton+10% Blocking Serum + Primary Antibody)/PBS (overnight, 4oC) (ratio 1:500)
5. Secondary antibody: Remove Primary antibody, wash 4 times with PBS --> Incubate cells in working solution of (0.1% Triton+10% Blocking Serum + Cy3-conjugated-Secondary Antibody (ratio 1:500) + DAPI (ratio 1:1000))
--> Washing 3 times with PBS

Good evening (at least in me time-zone)
I have been having some trouble with some routine Immunological staining. What the ideal conditions for IHC slides before staining is? Should I leave these slides to drain overnight at room temperature or use the slides dryer for 3 hours at 45 degrees? By the way, What is the appropriate temperature for a tissue flotation water bath?
I hope to receive any information, suggestions, leads, comments, questions, and/or thoughts you may have.
Thank you for your time and advice!
Sam
I have isolated PBMC from human blood and have set up an MLR for 5 days. I have then stained with CFSE and the cells have not proliferated.
Any help would be appreciated
Peroxidase activity
📷📷
Dormant seed Non dormant seed
I have a question about some weird nuclei I see while imaging. I'm sorry if this isn't the best place to write it!
One image has bad, "shattered"-looking nuclei from dentate gyrus.
The other image has healthy nuclei from CA1.
I end up in some experiments with really terrible looking DAPI with "shattered" nuclei. Sections with this type of DAPI are highly correlated with really bad FISH staining as well. It seems like most of the tissue integrity is lost in general.
I've seen this before, but not enough times to know what step of tissue collection and processing could cause it. I don't *think* this is purely a cryosectioning issue.
Does anyone have any guesses for what could cause this issue? I'm guessing I can't be the first person to run into this!
Tissue processing
I am imaging coronal brain sections from mouse tissue that is flash frozen immediately after dissection. In this case this is imaging dentate gyrus, where nuclei should be exceptionally dense. 20 um cryosections are immediately placed onto Superfrost glass slides and stored at -70 (in this case for ~2 weeks).
Sections are fixed with 4% PFA on the slide and then dehydrated with serial EtOH incubations of 50%, 70%, and 100% for 5 minutes each. Then sections go through the RNAscope smFISH protocol, at the end of which they are stained with DAPI for 30 seconds.


I want to stain bull sperm cells (dead/alive) with Hoechst 33342 (10 mg/mL in H2O) and don't know how to do it properly. I will be grateful if you could help me. Best regards and stay healthy.
We have a demarcation and analysis protocol in ImageJ for images of C. elegans stained with Oil Red O. However, it does not seem to be the best way and we were unable to find an easy-to-execute protocol.
We have constructed lentivirus transduced U251 cell lines to express (1) EGFRvIII (its amino acid sequence is MRPSGTAGAAFLALLAALCPASRALEEKKGNYVVTDHG..., which is identical to the sequence described in https://www.addgene.org/20737/)-mCherry or (2) EGFRvIII_G38C(MRPSGTAGAAFLALLAALCPASRALEEKKGNYVVTDHC...)-mCherry.
Both types of cells did not yield positive results by using antibodies against LEEKKGNYVVTDHC (https://www.novusbio.com/products/egfr-antibody-dh83_nbp2-50599af647).
And the (1) EGFRvIII-mCherry U251 cells were also tested negative using 2-step staining (https://www.emdmillipore.com/US/en/product/Anti-EGFRvIII-Antibody-clone-DH8.3,MM_NF-MABS1915-25UG + https://www.thermofisher.cn/cn/zh/antibody/product/Donkey-anti-Mouse-IgG-H-L-Highly-Cross-Adsorbed-Secondary-Antibody-Polyclonal/A32766),
The (2) EGFRvIII_G38C-U251 cells have not been tested by 2-step staining.
We are curious about this question. Have you met problems when dealing with EGFRvIII?
Hi, I have used masson's trichrome staining and trying to analyze collagen staining.. How can we analyze it using Image J. Anyone have the macros available?
Thanks
I have already carried out the lipid staining test in C. elegans with oil red, however, in the last tests I am unable to stain all the animals efficiently, most of them do not stain or only have part of the lipid droplets stained.
I don't know where I'm going wrong in the protocol, maybe when preparing the 0.3% oil red solution in 60% isopropanol.
I have 5 primary antibodies for immunofluorescence (IF) Staining of tissues. For that, I have only FITC conjugated secondry antibodies. If I'll do IF for 5 different targets but all with same secondary antibodies (FITC), will that create issue in publishing? Will the editor/reviewers ask that it's confusing if the authors have really used 5 different antibodies for IF or they just used one type of anitbody Staining, and they are just showing different parts of same tissue to be considered as 5 different types of antibody IF Staining!!!
Hello,
I do IP using HA-tagged beads. One of my proteins through which I precipitate complex has HA-tag. I see this protein (with HA-tag) after IP. The signal is strong and bright.
The complex contains several proteins. For each protein I do separate western blot (I have different membranes for different proteins - I do not do re-probing). Proteins without HA-tag are not very abundant in the precipitated complex and I need use Clarity max (Femto) staining to see them. As a result I see as proteins that I try to detect as HA-tagged protein. The signal from HA-tagged protein is weak but it is still crucial for me because one of proteins that I am looking for has size slightly bigger than HA-tagged protein.
I tried to use true blot secondary antibody but it did not helped. I tried to use different primary and secondary antibodies, for example antibody fro HA-tagged protein is produced in mouse and I use anti-rabbit primary and secondary antibody for other protein of my interest but it did not helped.
I do blocking of membrane in 5% milk with tween-20. Primary and secondary antibodies are also diluted in milk. As an option I tried to block membrane and incubate with antibodies in 5% milk with TBS-T and wash it with TBST but it did not helped.
I cannot increase number of washes because I will loose proteins without HA-tag.
Does somebody has an idea how to solve this problem?
This is the first time I did an H&E staining, and I found that there are some white (blank) spaces between the cells. I do not think this is the normal morphology of this tissue.
Here are two screenshots of the tissue. They are from exactly the same microscope slide, both of them are mouse heart tissue, and I think most of the cells in the picture are cardiomyocytes. They are 2 consecutive slices from a frozen cryotome.
My own opinion is:
1. I don't think these are adipose tissue.
2. In both screenshots the longitudinal muscle (top part) looks alright. The transverse muscle (bottom part) in picture 1 has white spaces, while the transverse muscle (bottom part) in picture 2 is normal. I am wondering if it is because some of my operations has damaged the tissue? and the transverse muscle is more prone to the damage(?)
This is the protocol that I followed:
1. Do fresh frozen sectioning;
2. Submerge the slide in 100% MeOH at -20 degree celcius for 30 min;
3. Remove slide, let MeOH evaporate;
4. Stain with hematoxylin for 3 min, wash by dipping in beaker with water for 15 times. Repeat with another beaker of water.
5. Stain with buffered eosin for 1 min, wash by dipping in beaker with water for 15 times. Repeat with another beaker of water.
6. Airdry.
Could anyone please tell me what might be wrong?


I would like to stain the suspension cells with DAPI to examine under the fluorescence microscope. kindly suggest me a protocol for the same.
I've been trying to study cell cycle using PI (propidium iodide + Triton-X + sodium citrate)b staining solution.
I trypsinize my cells (A431: skin carcinoma), fix them using 100% methanol and store in 4 degree Celsius. I then wash the cells once with cold PBS and re-suspend them in the staining solution. Till this point it is a single cell suspension after vortexing well.
I leave the samples overnight in 4 degree Celsius and acquire using low cytometer the next day. But by this time the cells form clumps. These clumps are not broken down by vortexing or even by pipetting.
I've tried vortexing the sample again and again right before acquiring the cells. I've used freshly made PBS and PI staining solution. I've maintained adding cold solutions since every step after trypsinization. I've shifted to using methanol instead of ethanol for fixation.
I'm still unable to solve for the clump formation.
"Hello,
I am a Ph.D. student at the Gwangju Institute of Science and Technology in South Korea, and I have some questions regarding my immunofluorescence (IF) experiment.
1.High background:
I have noticed that my IF image exhibits a high background, as there is significant staining in the broader areas of the cell. I suspect this may be due to various factors such as high concentration of Ab and problem about secondary Ab.
2. Non-specific staining:
I have observed staining in conditions where cells are not expected to be stained, which leads me to believe that there may be non-specific staining occurring. Is this stained? I am unsure if my troubleshooting approach is correct in addressing this issue.
As this is my first time conducting an IF experiment, I would greatly appreciate any guidance or advice on how to address these challenges effectively.
Thank you.


Hi, I just want to ask regarding flow cytometry method for indirect staining of U87 cells (Glioblastoma) as I'm still quite new in using this method. My objective is to observe the expression of CD133 from U87 cells.
For the control samples, I used Unstained and Isotype control. Right now, I'm facing a problem where my Unstained result is same as Isotype and my stained samples (which is positive samples). The histogram result show that my unstained, isotype and stained have same peak even though I've already tried adjusting the voltage but the result is still the same. Is there any recommended solution for this problem? Also I want to know how do we gate the negative and positive cells population properly by using unstained control as negative? Thank you and looking forward your positive responses.
For primary antibody, I'm using CD133, meanwhile for secondary is Alexa-fluor@488 conjugated. I didn't use any blocking agent. For FACS buffer, I'm using 5% FBS/PBS.
I usually fix cells with Formaldehyde 3.6-4% at 4 degrees, I was having a poor staining, may this be due to the crosslinking induced by formalin? additionally, what is the consequence if reducing the wasing after incubating with formalin?
Most staining protocols for flow cytometry in 96-well plates use V-shape or U-shape plates. I would like to ask if staining could also be done in flat-bottom plates.
Thank you!
Most of the Antibody company (having pr. Ab conc. range 0.2 mg/ml - 0.5mg/ml) websites suggests dilution around 1:200, but it seems not staining or faintly staining. What is the hand on experience on bench for scientists performing IHC/ICC?
Hi, I have grown primary nasal cells on semi-permeable trans-well (PET) inserts and would like to prepare a slide (for confocal microscopy). I imagine it has to be fixed and cut out and placed on the glass slide. Does anyone know how to fixate it on the slide without it moving around so its possible to stain it ?
Your help is much appreciated.
Thnak you!
I have a question for the professionals. The essence of the problem: I fix the oligos on a plastic substrate, they play role of primers in solid-phase PCR. I carry out a one-step PCR with simultaneous labeling of the product with biotin (I add 10% labeled uridine to unlabeled T to the DNTP mixture), then I wash it in PBS and incubate it with the streptavidin-peroxidase complex and then incubate it with the substrate for peroxidase. Everything would be fine, but in the control wells, where the PCR reaction mixturedoes not contain DNA, I have a staining of oligo spots, weaker than in the experimental wells, but it is there. Moreover, in the control wells, where only PBS was added, weak staining also appears at the localization spots of the oligos. If I simply add the complex to the wells (without any PCR treatmen), then only the positive control points, that is, the initially labeled oliagos, are stained in them. So here's the question. Can streptavidin (or peroxidase) bind to something other than biotin or DNA oligos? I’ve been fighting with the problem for a couple of months now. I 've changed blocking buffers, polymerases, washing modes, but the result is still the same. Help, good people!
Hi! We are using DAB to stain biocytin filled (20-40min) interneurons in spinal cord. As shown in the figure, the soma is stained well, but not the dendrites. Could someone tell me what might be the reasons? Thank you!

For instance, using one hemisphere for brain slice staining via IHC and the other for analyzing neurotransmitter levels through LC-MS/MS? Are there any existing references or studies demonstrating this combined approach?
Hello Good people
When I stained my adherent non-transfected cells with Hoechst 33342 staining it showed blue fluorescence but dull staining happened with GFP transfected cell
I used 2ug per molar
30 min incubation at RT
300ul per well in 12 wells plate
So, what's your suggestion for better procedure to be able to see cell segmentation more clearly!
What's the benefits from PBS washing as recommended by some protocols at the beginning or the end!


We have an experiment that will look at the impacts of treatment on the proliferation of total muscle fibre (i.e. combining primary, secondary and tertiary) in the skeletal muscle of pigs. Technically, the total muscle fibre number count is usually conducted using muscle tissue section staining (eg, nuclei stain or specific antibody), which requires either biopsy or euthanasia of the experimental animals. To avoid this invasive sampling procedure and to achieve better animal welfare, are there any circulating biomarkers (with/without challenge) that can be used as an estimation of total muscle number (e.g., the circulating biomarker is correlated with the total number of skeletal myofibre)?
Thank you
Kind Regards,
Fan
I am wondering if there are any commercially available fluorescent stains for live imaging of astrocytes derived from hiPSCs. We are co-culturing neurons and astrocytes and would like to stain the live cells to determine the change in their populations over time.
I want to observe chemotactic movement of e coli using fluorescence microscope. DAPI staining protocol didn't work for me and syto 9 is too expensive.
Hello all!
I have som trouble staining beta galactose in my MC3T3 cells.
Here is my protocol:
- Cells stressed with H202 treatment.
- washed twice in PBS
- fixated in 4% paraformaldehyde 5min at 4 degrees.
- washed 3times ×3min in PBS
- staining solution added (freshly made. Containing 1mg/ml X-gal, 150mM NaCl, 40mM sodium phosphate and Citric acid, 5mM potassium ferrocyanide and potassium ferricyanide, 2mM MgCl2, pH 6)
-incubate over night, 37degrees in a non CO2 chamber
What is missing or could be optimised for the staining to work?
Grateful for any help!
Hello,
I'm conducting a beta galactosidase staining assay for my fibroblasts using X-Gal. I dissolve X-Gal (stock 50mg/ml prepared in DMSO) in the staining buffer at a final concentration of 1mg/ml. However, even after 24-48 hours of incubation at 37°C (w/o CO2), I'm unable to observe any staining. Additionally, X-Gal precipitates in the dish with prolonged incubation, forming crystals as shown in the picture.
I have also tried heating the staining solution to 65°C before adding X-Gal, but nothing seems to work. Kindly help me resolve this issue.
Composition of my staining solution: 5mM potassium ferrocyanide, 5mM potassium ferricyanide, and 2mM MgCl2 in 1x PBS (pH 6).
For fixation, I use 4% PFA in 1x PBS for 5-10 minutes followed by PBS washes twice before adding the staining solution with x-gal.

In the picture derived from light microscopy, there are some red stains apart from the blue ones. I am wondering, what could it be?

I'm planning an experiment where I can access intracellular cytokines in a specific subregion of the brain in mice. However, this brain region is quite small, maybe 50,000 cells per animal. I know I will need to pool mice but how many would I need to pool? Can I use 500,000 cells? Pooling more than 10 mice wouldn't be feasible.
Thank you!
I am currently conducting experiments involving SA beta-gal staining to detect senescent cells in my samples. I need guidance on the photography and cell counting aspects of my methodology. Specifically, I'm unsure about the best practices for photographing stained cells and determining the number of photographs needed per sample or experimental condition. Additionally, I'm seeking advice on the optimal approach for cell counting after staining, including the number of cells to count per field and the appropriate number of fields to photograph for reliable data analysis. Any insights or recommendations from researchers experienced in SA beta-gal staining and cell counting would be greatly appreciated. Thank you for your assistance!
Hello all,
I've been struggling to get good FAP staining on my tissues using fluorescent IHC. I've tried three different antibodies from different companies, but the staining isn't working well. Has anyone used an anti-FAP antibody for human cancer tissues and gotten good results confirmed by a pathologist? Any advice would be helpful. Thanks!
Hello,
My E. coli cells express both green fluorescent protein as well as mCherry. So I need a fluorescent stain of color other than green and red fluorescence to enumerate their viability. Please suggest. Thanks in advance.
I have been studying about Vagus nerve and I want to check the activity of the Vagus nerve in my research. Could you advice me about it?
So I am trying to live-image the synapses of NK cells with either A549 or K562 cells. For that I am going to stain NK cells with lysotracker, cancer cells with membrane stain, and then add a dead cell stain in the medium to visualize the real-time killing of cancer cells by NK cells.
In regards to K562 cells, which are suspension cells, I found a protocol that uses an antibody (mouse-antihuman glycophorin A) to bind K562 to the wells.
My question is, for folks that have done live-cell imaging of NK cells with any other suspension cell, what other type of surface coating could work besides that antibody?
Thanks in advance.
Hello everyone,
I am trying to do surface staining of a protein of interest in adherent cells for analysis in FACS. However, I am not getting what I am expecting, and I am wondering if something in my cell preparation is going wrong. Specifically, if I'm correctly treating the cells with the drugs. I would appreciate it if you could take a look at my current protocol and give some feedback if you think I'm missing/doing something wrong.
Here's my protocol so far:
1. Treat the cells for the desired time with the desired drug on the T-25 plates (cells have grown to the desired density for flow (~80% conf)).
2.Trypsinize the cells and spin down (at 4C) to remove trypsin ( I have transferred them to Eppendorf tubes)
3. Wash once with PBS (at4C)
4. Wash with cold PBS (at 4 C)
5. Add the primary antibody to each of the tubes and incubate on ice for 30 minutes.
6. Wash twice with Flow cytometry staining buffer from eBioscience (https://www.thermofisher.com/order/catalog/product/00-4222-26)
7. Add 3.7% PFA to fix cells at room temperature for 10 mins
8. Spin down to remove excess PFA
9. Wash with FC staining buffer
10. Resuspend in FC buffer for storage until FACS experiment. Store at 4C covering them with foil.
It is important to note that, starting from step 4, I have placed my samples on ice the whole time to prevent endocytosis.
Please let me know if you have any suggestions.
Thank you in advance,
Valeria
Does anyone know any protocol I can follow for muscle fiber-type staining for paraffin-embedded tissue?
I use 1ul SYTO9 and 1 ul PI per ml of water. The bacteria are attached to a surface and I cover them with 20 ul of this mixture for 10 mins (in the dark), before removing the dye and imaging. The dyes are mixed just before use. At 10 mins, MRSA on steel surfaces are staining both green(live) and red(dead), but I know by culture that they can survive on steel for hours if not days at a time. The filters do not allow cross-fluorescence. The culture is an O/N growth of MRSA in LB broth, and I centrifuge and re-suspend in PBS before use. I have reduced the amount of PI, I have washed the cells to try and remove extracellular DNA/media debris. I can't think of what else to do. Thanks in advance for any suggestions from people who also use this staining kit.
Which fluorescence markers are best to use for staining macrophages. I want to prepare sample to get training with the microscope for my research.
Hi everyone,
I performed an immunofluorescence (IF) staining for Ki67 using a validated, specific Ki67 antibody (Dako) and I see a clear upregulation of the protein expression compared to my baseline samples. However, on RT-qPCR, Ki67 is downregulated. Is this possible or should I question my staining (although the IF signal seems very specific to me)?
Thank you for your help!
Sara
I found the optimal antibody dilution and incubation time to stain cells of fish brain sections (50μm thick).
However I need to stain those cells in thicker brain sections and I was wondering what are the criteria to apply, if any, for the antibody dilution and incubation time so I can get results comparable to thinner section staining (i.e. Increasing the incubation time according to the thickness).
Looking forward to your feedback.
Dear colleagues, help me figure this out.
We are trying to analyze and sort antigen-specific cells using Flex T technology (Biolegend). After UV exchange, the efficiency is 60%. Next, according to the manufacturer’s protocol, we separate and carry out conjugation with streptavidin-PE and steptavidin-APC. And when using these reagents, we obtain a high level of nonspecific staining for each of the fluorochromes.
I can’t figure out why there could be such pronounced non-specificity and how to deal with it. I would be grateful for any suggestions)

Hi
Does anyone know which dye can be used for staining exosome membranes, aside from PKH67?
Thank you in advance for your help
I am using mouse serum to block Fc receptors before staining, for flow cytometry. My question is if I should also add the serum to compensation beads before staining them, so that the cells and the beads go through the same processing.
Thank you very much in advance!
I've always used abd as load control for western blot normalization, and use Ponceau S stain as a transference control (along with keeping an image - a picture taken by my cellphone - for supplementary paper material submission). My question is:
Can I use this picture to quantify total protein? Or there is a specific way/equipment to image the membrane stained with Ponceau S to quantify total protein?
how to count metastatic nodule in slides stained with H&E. different sections have different number of nodules. Should I add all or should I report the section that showed the highest number?
I am using it for intracellular staining. Why does it have to be made fresh every time?
I need to do fluorescent microscopy using Propidium Iodide. I initially fixed my cells with 4% paraformaldehyde and saw red stain in Control cells. It turns out PFA is cell permeable. So if anyone has a protocol using Ethanol as a fixative please do share.
I recently conducted staining on brain sections of adult zebrafish using Nissl stain. The brains underwent pre-fixation in 4% paraformaldehyde, followed by storage in 75% ethanol at -20°C for a period of time, before being rapidly frozen in methylbutane on dry ice in an OCT mold. Cutting was then performed at a thickness of 20 microns at a temperature of -15°C, using charged slides. The stained slides were mounted with DPX and left to dry at room temperature for three days.
Unfortunately, upon examination at 10X magnification, not the entire slice is in focus. I also attempted to use gelatin-treated slides instead of charged ones, which yielded only slightly improved results.
I suspect that these issues may be attributed to two factors: 1) inadequate adhesion of the brain to the slide due to insufficient stickiness of the slide, and 2) the formation of micro-bubbles between the slide and the slice during the cutting process.
Please share your experiences or suggestions regarding this matter. Thank you!

Hello !
For my microbiology project i need to visualize living P.lunula under a lightmicroscope.
I saw that you can try using Toluidine blue stain, but have not found much research about it.
Hi,
I could see this morphology under the microscope (black arrow, 20x). Could someone identify what this morphology/ structure could be? and if so how to stain it with a specific dye?
Cells are colorectal cancer cell line, this structure does not stain with DAPI.
Thanks!

I have tried 4 IBA1 antibodies and cannot seem to get good staining. Mice were perfused/fixed with PBS/4%PFA. Brains were placed in PFA overnight, and then moved to 30% sucrose and frozen in OCT after sinking. My sections typically tend to be thicker (30um or 35um), but we do sections on slides instead of free-floating due to less handling and integrity of the structures. All of my antibodies work except for these IBA1 antibodies. I have tried permeabilizing with triton and saponin and got similar results. (Fix for 5 minutes on slide with 2% PFA, perm with 0.3% triton 15 min, block with 10% goat serum 30 minutes, then primary and secondary incubations with blocking serum). Antibodies are spun before addition.
Can anyone advise me as to why these IBA1 antibodies are creating so much background at both 1:100 and 1:1000, and why it is not staining the filaments of the microglia? Any advice is greatly appreciated.
I recently conducted staining on brain sections of adult zebrafish using Nissl stain. The brains underwent pre-fixation in 4% paraformaldehyde, followed by storage in 75% ethanol at -20°C for a period of time, before being rapidly frozen in methylbutane on dry ice in an OCT mold. Cutting was then performed at a thickness of 20 microns at a temperature of -15°C, using charged slides. The stained slides were mounted with DPX and left to dry at room temperature for three days.
Unfortunately, upon examination at 10X magnification, not the entire slice is in focus. I also attempted to use gelatin-treated slides instead of charged ones, which yielded only slightly improved results.
I suspect that these issues may be attributed to two factors: 1) inadequate adhesion of the brain to the slide due to insufficient stickiness of the slide, and 2) the formation of micro-bubbles between the slide and the slice during the cutting process.
Please share your experiences or suggestions regarding this matter. Thank you!
UPD
I finally found a pattern for these unfocused brain areas - they stem from microbubbles formed during sectioning, which I cannot avoid, unfortunately. In the worst situation, when the bubble covers almost the entire area of the slice, the slices are washed off from the slide. In better cases, I observe the unfocused areas (please see the picture).


I've been testing a protocol to evaluate stained whole blood in a wet mount for a WBC differential using Toluidine Blue, which results in clumping.
The protocol requires a 1:50 ratio of whole blood mixed with normal saline, then a 1:1 ration of blood dilution to stain incubated at 37C for 5 minutes. 10ul are placed into a disposable hemocytometer for inspection.
The staining is perfect however the cells clump together creating difficulties in count and differentiation.
A blood smear is not an option in this particular scenario. Alternative stains for a WBC differential using a wet mount could be an option.
Any guidance would be appreciated. Thank you.
I'm attempting to stain cancer cells in suspension from a 12-well plate for my research project. Could anyone provide guidance on the most effective staining protocols and techniques for ensuring accurate and reliable results? Any insights or recommendations on suitable staining dyes, concentrations, fixation methods, and imaging procedures would be greatly appreciated. Thank you in advance for your assistance!
Has anyone extracted total RNA from stained H&E slides? We have cases in our study that have no tissue left in the FFPE blocks, and no other tumor source.
Hi everyone,
I established a murine spheroid for glioblastoma, and I want to fix and stain them to conduct immunofluorescences. I have tried several times, but the size of my spheroids are small, and I cannot see them during Paraffin embedding step. I would be grateful if you could give me some hints on doing this step.
After I treated the subcutaneous xenograft with an antibody conjugated to MMAE, the volume was smaller than that of the control group, but the Ki67 staining was enhanced. MMAE works by arresting the cell cycle at the G2/M phase.
We are in a pharma proces, and after and/or before the centrifugation, it's appearing a fresh mesh, and we hypothesize that is a lipid mesh. For identify it I thougth to use Oli Red O, but I'm not sure if it can be used to stain lipids of a suspension sample from tissue. There is another cheap method to identify it?
Tank you so much.
hi
i started to stain the umbilical cord vein for ephB4 using anti-rabbit primary and FITC
and I have signal in both control and samples. what could be the reason if anyone been through this?
note: I stained for different marker(not ephB4) and I got similar signal in both control and sample
I followed an immunofluorescence staining protocol for my cells differentiated into neurons at D14. Using 4% PFA (Alfa Aesar product) for fixation in PBS, I observed that 2-3 out of 8 wells were empty or neurons were drift towards one side of the well after D1. Upon completing the protocol and examining under a fluorescence microscope, I noticed 7 out of 8 wells were empty, with one exhibiting perfect fixation and staining. What might be causing this issue?
Can you help me with the calculations? I am performing crystal violet staining protocol.
In the anatomical and microscopic observation of the leaf and stem of the plant, can I first take a sample and then proceed to cross-sectioning, staining, and microscopic observation in the following days?
Hi:
I am performing immunofluorescence staining on 100 um 4%PFA fixed pancreas sections using our self-made polyclonal primary antibody. Tissue was gradient dehydrated in methanol diluted in 0.2% NP40. The sections were blocked with 1% BSA, 4% FBS, and 0.1% Tween-20 in PBS for 1 hr. But end up with a poor staining result under confocal examination. Is there any possible solution? Thanks!
Hi,
I had stained CD40 antibody on tonsil paraffin sections. But staining came positive only on periphery of tissue and there is no staining in the cente of tissue. What can be the reason??
I am working on a small protein (15 KD) ; their is no information about its function ;
we can't see it in western blot and we stain it but we cant see in the gel (or maybe can see but we sisnt got that)
Hi, I am new to this platform. I am currently preparing for Western Blot experiments using Young (7-10 mo) and Aging (20+ mo) mice and the protein of interest will be tight junction proteins (ex. occluden, claudin-5, ZO-1).
I have looked through a series of commonly used housekeeping genes (e.g. β-actin, α-tubulin, β-tubulin and GAPDH) but found that all of them will be affected by aging. I have also looked through stain-free and total protein normalization (by Ponceau S or Coomassie staining), which seems to give a more promising result than the housekeeping genes. But since they are relatively new approaches, I would like to seek opinions here about a good way for the loading control of age-related Western Blot.
Thank you very much!
I was performing IHC on paraffin embedded mice vascular tissue ,after examining the sections under microscope,I found irregular transparent spots on top of my tissue section, please help,thank you.

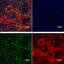
I stained various markers (CD3, CD8, CLEC4F, etc) in liver tissue containing metastatic tumor nodules for immunofluorescence imaging, but when I tried to take a picture showing tumor region and liver region together in one frame, there were too many non-specific background fluorescence in liver region. I tried adjusting the fluorescence to get rid of the background staining, but adjusting it based on liver tissue made positive staining in tumor region fade away. (I attached an image for your reference) There was no such problem when I stained the tissue with TUNEL and DAPI, which both stain DNA.
It seemed like autofluoresence and non-specific binding could be the problem, so I am trying to redo the experiment in perfused liver tissue (containing metastatic tumor nodules) and also change blocking solution (From 5% BSA + 0.3% Triton X-100 in PBS to 1% BSA + 5% Normal serum + Glycine + 0.3% Triton X-100 in PBS, RT for 2 hours).
I was wondering if anyone else has also experienced the same problem when staining liver tissue for IF imaging. If so, could you please share how you handled the problem?
Thank you!
I'm facing a puzzling question. I stained two different cell lines of the same type and observed DAPI dots in the cytoplasm of one, unlike the typical DAPI pattern in the other (see images). Both were untreated and on the same plate, subjected to similar treatment.
Has anyone encountered a similar issue? Any hypothesis of what can it be? We have done mutiple mycoplasma testing by PCR and turned out negative. Also, if contamination is present and the cells share the same plate, shouldn't the contamination transfer between them?
thank you very much for your help



I subcutaneously performed implantation in rats and harvested the sample after 1, 2, 3, and 6 months. The implant is decellularized human skin. The procedure of how my lab did it was drawn in the attached picture.
Then I carried out IF staining with alpha-SMA, and Vimentin for the sample to monitor fibroblast migration into the implant. The magnification of the picture was 20X, therefore, the region that appeared in the picture is the implant area. I observed cell morphology and also something that looks like a "thread"(?), a very thin structure and don't look like cells at all. I'm not sure what that is and I tried to find references but I could not find anyone who did the same staining for human skin implant.
Could someone please help me with this problem? Please let me know if you have any opinion on this. Thank you so much




I want to embed a 2 mm thick tissue sample (with a polymerized diaminobenzidine stain) into glycol methacrylate and use synchrotron phase contrast x-ray microtomography to image the tissue. However, I am not sure whether glycol methacrylate is translucent to x-rays at wavelengths ranging from 15 to 30 keV. Could someone please let me know if they have ever had success with imaging through this material via x-rays?
In addition, I was wondering if the x-ray phase decrement for polymerized diaminobenzidine is distinct enough from that of glycol methacrylate plastic that it should give good contrast. Thanks!
Hi,
Been looking for the method to stain astrocytes using the Cajal's gold sublimate technique.
Anyone knows?
Thanks
Hello, I have a problem with staining the mouse brain.
These days, my professor told me
"There is something wrong with your images. I think you must have made the mistake in perfusion or fixation. It's not compact. There are too many holes"
But I can't fix the problem.
Could you help me, please?
There are vacancies around the nucleus like edema in H&E and IF images.
I think it is just a nucleus hole.


Hello everybody. We have a problem with western blot.
wet transfer 100 min, 75mA in TGS 1x + 20% metOH.
does anyone help us to rescue this problem?
in attached you can see the ponceau staining image.
thanks

Hi flow users/cell masters,
I'm just wondering if anyone here has experience with staining cytoplasmic cytokines after staining extracellular proteins. So basically what I did in my protocol was staining extracellular markers using PE-conjugated and APC/Cy7-conjugated antibodies, then fix/perm my samples using the 1X fix/perm buffer eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set before staining cytoplasmic cytokine using antibodies resuspended in 1X perm buffer.
I didn't see any APC/Cy7+ or PE+ populations in flow, which is weird coz the markers are supposed to be highly expressed. I saw some posts on Reddit that APC/Cy7 is not stable, but I don't know if PE is unstable either. Also, does anybody know if eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set contains methanol? I couldn't find the info anywhere, if it is, it makes sense then... Since PE and APC tandems are not methanol-resistant.
Thanks in advance to anyone who's going to answer this :)
I have a problem and am asking for advice.I am doing WB. For electrophoresis
I use an 8% separating gel and a 4% thickening gel. The electrophoresis has 2 phases : 30 min-90V , 60 min 110V. Electrophoresis buffer from Bio-Rad of composition 10xTris/Glycine/SDS. For transfer I use Bio-Rad's ready-made but diluted Transfer Buffer. I perform a standard transfer to PVDF membranes (30 min). Membranes are incubated with milk and with tris pH=7.6 and tween. Then as primary antibody I use B act polyclonal antibody from Invitrogen, Lot YD371542, as secondary antibody anti-rabbit IgG HRP Conjugated HAF 008 from R&D, Lot FIN 1922041. Then I use Precision Protein StrepTactin-HRP Conjugated 5,000x. And I add calling reagents.
Why does my 70 kDA stain very clearly and my beta actin stain very weakly ( 42-46 kDa) ?
I make WB from homogenised cardiac tissue. To prepare it, I used Thermo Scientific protease inhibitor at 225 microlitres per 25 ml homogenization buffer.
I attach a blot of beta actin below .
How can we explain the fact that a LAB stain that resists to low pH (2) becomes sensitive after 2 years of conservation at -20°C ? Notice that some problems of power cut occured during this time lapse.
Hi all, I have been trying to detect several polypeptides by WB. And tricine-sds-page (16.5% or 20%) and glycine-sds page(20%) gel electrophoresis followed by Comassie blue staining showed no bands below 7.8kda marker, either bands in blotting(antibody works well). The attachment file is the Coomassie blue staining of a 20% tricine gel.
Any experience or communication will be appreciated!

Purpose of using stain and de-stain in SDS-PAGE gel
can you provide me with the best p16/ki67 dual staining protocol for FFPE tissue (cervical)? and what are the best antibody brand for that?
Hello everybody. We have a problem with western blot.
wet transfer 100 min, 75mA in TGS 1x + 20% metOH.
does anyone help us to rescue this problem?
in attached you can see the ponceau staining image.
thanks

Hi, I am trying to stain OP9 cells with ALP antybody for FACS. I tried 2 different secondary antibodies with no success. I get a signal from the secondary antibody alone. It's like the second antibody can somehow connect to the cell. Did anyone notice this problem?
Thanks
Hi. We are trying to find information about Trihalo compound so we can make our own stain-free gel in the lab rather than having to buy it.
Has anyone tried it already? What is the make of Trihalo compound? I can only find Trihalomethanes to buy but not sure if they are the same thing. Found a few articles talking about adding 2,2,2-trichloroethanol in polyacrylamide gels.
Any suggestions are very welcome!